PeptideShaker Database Requirements (0.22.4 or earlier)
PeptideShaker likes specific format for database if you don’t use searchGUI. They recommend Uniprot database. But occasionally I want to use my own database. There are two specific requirements for correct database in PeptideShaker.
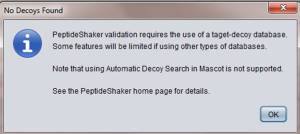
1) It needs decoy sequences.
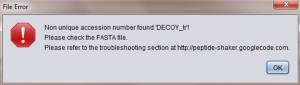
2)Accession number for decoy sequence has “_REVERSED” tag.
For 1), if you don’t contain decoy sequence, it complains “No Decoys Found”.
For 2), if the tag is not correctly added, no unique accession error appears.
So the correct fasta format for PeptideShaker is
>IPI000123456.7:protein_description
MEHKEVVLLLLLFLKSGQGEPLDDYVNTQGASLFSVTKKQLGAGSIEECAAKC
EEDEEFTCRAFQYHSKEQQCVIMAENRKSSIII
>IPI000123456.7_REVERSED:protein_description
MEHKEVVLLLLLFLKSGQGEPLDDYVNTQGASLFSVTKKQLGAGSIEECAAKC
EEDEEFTCRAFQYHSKEQQCVIMAENRKSSIII
These requirements are a little bit problematic. I also use IDPicker for MS/MS spectra analysis and it uses decoy tag before the accession number. Well, all I have to do is to create two databases and search with different databases for IDPicker and PeptideShaker.
If you are using searchGUI to search your MS/MS spectrum with OMSSA & X!tandem, you don’t have to worry about these requirements. The software will take care of them.
A minor precision:
By default, PeptideShaker recognizes as decoy all proteins starting or ending with “REVERSED”, “RND” or “SHUFFLED”. In the newest version you can set your own tag 🙂